By Daniel Dunaief
Mirna Kheir Gouda arrived in Commack from Cairo, Egypt, in 2012, when she was entering her junior year of high school. She dealt with many of the challenges of her junior year, including taking the Scholastic Aptitude Test, preparing for college and adjusting to life in the United States.
Her high school counselor at Commack High School, Christine Natali, suggested she apply to Stony Brook University. Once she gained admission, she commuted by train to classes, where she planned to major in biology on the road to becoming a doctor.
She did not know much about research and wanted to be involved in it to learn, especially because Stony Brook is so active in many fields.
“After some time conducting research, I came to be passionate about it and it was no longer just another piece of my resume, but rather, part of my career,” she explained in an email.
She reached out to Gábor Balázsi, a relatively new faculty member at the time, who suggested she consider joining a lab.
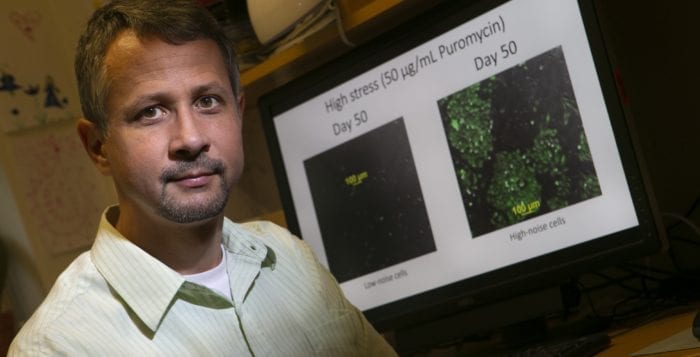
Balázsi uses synthetic gene circuits to develop a quantitative knowledge of biological processes such as cellular decision making and the survival and evolution of cell populations.
Balázsi knew Kheir Gouda from the 2015 international Genetically Engineered Machine team, which consisted of 14 members selected from 55 undergraduate students.
“Having this iGEM experience,” which included deciding on a project, raising funds, carrying out the project and preparing a report in nine months, was a “very promising indication” that Kheir Gouda would be an “excellent student,” Balázsi explained in an email.
Kheir Gouda chose Balázsi’s laboratory, where she worked with him and his former postdoctoral fellow Harold Bien, who offered her guidance, direction and encouragement.
As a part of the honors program, Kheir Gouda had to conduct an independent research project.
She wanted to “work on a project that involved adaptations and I always thought, ‘What happens when the environment changes? How do cells adapt?’”
She started her project by working with a mutant gene circuit that was not functioning at various levels, depending on the mutation. She wanted to know how cells adapt after beneficial but costly function loss.
An extension of this research, as she and Balázsi discussed, could involve a better understanding of the way bacterial infections become resistant to drugs, which threaten their survival.
“The idea for the research was hers,” Balázsi explained in an email. Under Bien’s mentorship skills, Kheir Gouda’s knowledge “developed quickly,” Balázsi said.
Balázsi said he and Kheir Gouda jointly designed every detail of this project.
Kheir Gouda set up experiments to test whether a yeast cell could overcome various mutations to an inducer, which regains the function of the genetic gene circuit.
Seven different mutations caused some type of loss of function of the inducible promoter of the gene circuit function. Some caused severe but not complete function loss, while others led to total function loss. Some were more able to “reactivate the circuit” rescuing its function, while others used an alternative pathway to acquire a resistance.
The presence of the resistance gene was necessary for cell survival, while the circuit induction was not necessary. At the end of the experiment, cells were resistant to the drug even in the absence of an inducer.
“This synthetic gene circuit in yeast cells can provide a model for the role of positive feedback regulation in drug resistance in yeast and other cell types,” Balázsi explained.
Kheir Gouda said she and Balázsi worked on the mathematical modeling toward the end of her research.
“What our work suggests is that slow growth can turn on quiescent genes if they are under positive feedback regulation within a gene network,” Balázsi wrote.
This mathematical model of limited cellular energy could also apply to cancer, which might slow its own growth to gain access to a mechanism that would aid its survival, Balázsi suggested.
Recently, Kheir Gouda, who graduated from Stony Brook in 2018, published a paper about her findings in the journal Proceedings of the National Academy of Sciences, which is a prestigious and high-profile journal for any scientist.
“Because PNAS has a lot of interdisciplinary research, we thought it would be a good fit,” Kheir Gouda said. The work she did combines evolutionary biology with applied math and synthetic biology.
The next steps in this research could be verifying how evolution restores the function of other synthetic gene circuits or the function of natural network modules in various cell types, Balázsi suggested.
Kheir Gouda’s experience proved positive for her and for Balázsi, who now has eight undergraduates working in his lab. “The experience of mentoring a successful undergraduate might help make me a better mentor for other undergraduates and for other graduate students or postdoctoral researchers, because it helps set goals based on a prior example,” Balázsi said.
He praised Kheir Gouda’s work, appreciating how she learned new techniques and methods while also collaborating with a postdoctoral fellow in Switzerland, Michael Mahart, who is an author on the paper.
“It is unusual for an undergraduate to see a research project all the way through to completion, including a publication in PNAS,” marveled Balázsi in an email. He said he was excited to have mentored a student of Kheir Gouda’s character.
Kheir Gouda has continued on a research path. After she graduated from Stony Brook, she worked for a year on cancer research in David Tuveson’s lab at Cold Spring Harbor Laboratory. She then transitioned to working at the Massachusetts Institute of Technology for Assistant Professor of Chemical Engineering Kate Galloway. Kheir Gouda, who started working at MIT in October, plans to continue contributing to Galloway’s effort until she starts a doctoral program next fall.
Kheir Gouda said her parents have been supportive throughout her education.
“I want to take this opportunity to thank them for all the sacrifices they made for me,” Kheir Gouda said.
She is also grateful for Balázsi’s help.
He has “always been a very supportive mentor,” she explained. She would like to build on a career in which she “hopes to answer basic biology questions but also build on research and clinical tools.”




 Understanding the basic science of how genes in individual cells respond to temperature differences could have broad applications. In agriculture, farmers might need to know how genes or gene circuits that provide resistance to a pathogen or drought tolerance react when the temperature rises or falls.
Understanding the basic science of how genes in individual cells respond to temperature differences could have broad applications. In agriculture, farmers might need to know how genes or gene circuits that provide resistance to a pathogen or drought tolerance react when the temperature rises or falls.




