An international scientific project that compares the genomes of 240 living species of mammals has identified transposable elements (TEs) – genes that can change their position within a genome, creating or reversing mutations and thus altering a cell’s genetic identity – as a crucial area of study to help uncover the evolutionary process of mammals and to better understand biodiversity. Stony Brook University’s Liliana M. Dávalos is a collaborator in the analyses of TEs for the project. Two new papers, one published in the current issue of Science, and the other in Molecular Biology and Evolution, highlight the findings.
The past 100 million years has caused mammals to adapt to virtually every environment on the planet. The Zoonomia Project, of which Dávalos is a scientific contributor, has cataloged the diversity in mammalian genomes by completing comparative genomic DNA sequences from the 240 species. The team, which consists of more than 150 scientists worldwide, published their multi-year comparative genome analysis in the Science paper.
Dávalos studies how biodiversity changes through time and what biological processes fuel biodiversity. She teamed up with David Ray and his lab at Texas Tech University to qualitatively analyze the dynamics of TEs.
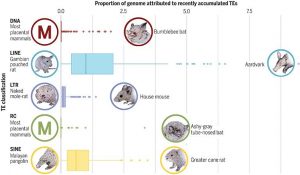
The paper describes the TE repertoires of 248 placental mammals. TEs make up a sizeable proportion of all mammalian genomes, yet there is much variation from one species to the next. The scientific team points out that that relating TEs to biodiversity is far from simple. Additionally, with the ability to move throughout the genome, TEs can contribute to biodiversity or also stymie it.
“Determining how many transposable elements of each kind are in each species is key to figuring out how transposons contribute to biodiversity. It seems simple to relate these counts to the number of species or their ecology, however that is misleading,” explains Dávalos, Professor of Conservation Biology in the Department of Ecology and Evolution, and a co-author of the paper. “Some species , like bats and whales, believe it or not are more closely related to each other than to others, such as bats and primates, so we must factor this related into our statistics within the comparative genomic mammalian analyses.”
The researchers identified more than 25,000 TE sequences in the mammalian set, with some mammals having large portions of TEs in their genome, calculated over time for each species. The average was approximately 45 percent
Overall, they concluded that “considering the wide-ranging effects that TEs impose on genomic architecture, these data are an important resource for future inquiries into mammalian genomics and evolution and suggest avenues for continued study of these important yet understudied genomic denizens.”
In the Molecular Biology and Evolution paper, novel statistical approaches to determining genome sequences in bats developed by Dávalos were used by the authors to describe the place bats hold with regard to TEs in mammals.
According to the lead author, Nicole Paulat, a graduate student in the Ray Lab, the research team found bats uniquely have more events involving TE transfers from one species to another. One mechanism that may explain such excess transfers is through viruses, an important finding on how several bat species have been found to host diverse and sometimes dangerous viruses.
Both papers based on the work from the Zoonomia Project illustrate that TEs are highly active across the genome of most mammal species, and because of this, future studies centering on TEs may help provide answers to mammalian biodiversity worldwide. Such research may also provide further hints as to how and why TEs disrupt mammalian genomes, therefore changing DNA and contributing to evolutionary processes and/or the development of disease.











