By Daniel Dunaief
They were in a terrible mood. They had spent an entire day searching for clues about creatures that walked the Earth millions of years ago and had come up empty.
“We were not finding even a single bone, nothing,” recalled Isaiah Nengo, who will be an associate director of the Turkana Basin Institute and an assistant research professor at Stony Brook University this fall.

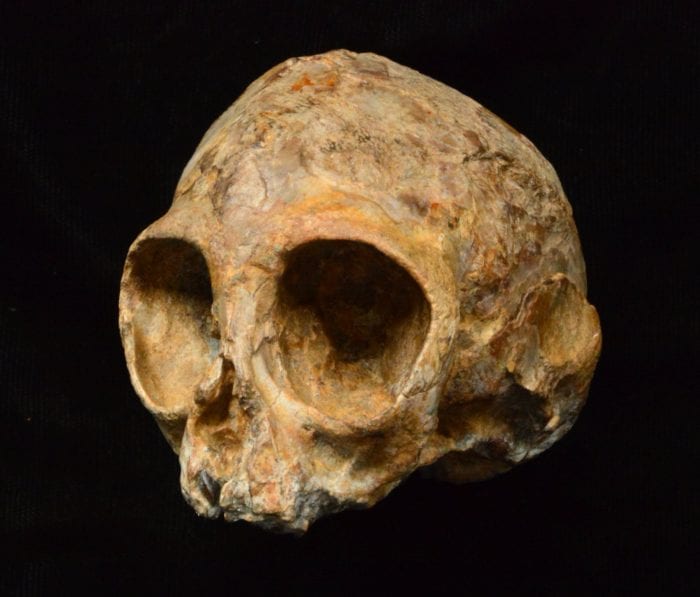
One of the fossil hunters in the group, John Ekusi, started rolling a cigarette. Nengo told him to move away from them so that they didn’t inhale second-hand smoke. Walking ahead, Ekusi made a spectacular discovery that Nengo called a “freak of a fossil.” Ekusi pointed out a bone sticking out of the ground that looked like the femur of a large animal. When they got closer, they could see that it had brow ridges. Pushing aside dirt, they saw the outline of a primate skull.
“We knew we had found something unique and we started celebrating right there,” Nengo said. “We were dancing and high-fiving. The thrill was unimaginable.”
Nengo and his team discovered the fossil on Sept. 4, 2014, in northern Kenya. This week, a team of researchers from the United States, France and England are unveiling three years worth of research into this remarkable find in the prestigious research journal Nature.
For starters, the researchers had to confirm the date of their fossil, which was about the size of a lemon. Rutgers University geologists Craig Feibel and Sara Mana studied the matrix around the fossil and the area around it.

“There was no doubt that [the fossil] came from this deposit and hadn’t rolled in or washed in” during some later period, explained Ellen Miller, a professor of physical anthropology at Wake Forest University.
Next, they had to figure out what kind of primate they had: It could have been an ape or a monkey. Fred Spoor, a paleontologist at University College London, did an initial CT reading using a medical scanner. He found intact molars that were characteristic of apes.
The researchers wanted to do a more thorough analysis of the three-dimensional shape of the skull, so they called Paul Tafforeau, a paleoanthropologist specialist of X-ray imaging who works as a beamline scientist at the European Synchrotron Radiation Facility in Grenoble, France. Typically, such research centers require scientists to wait a year or more.
As soon as Tafforeau saw the photos, Nengo recalls, he said, “You can bring it in anytime.” Tafforeau used a technique called propagation phase contrast–X-ray synchrotron microtomography. In an email, Tafforeau described it as being close to a medical scanner, but 1,000 times more precise and sensitive.
Over the course of three or four days, Tafforeau analyzed the teeth that hadn’t erupted from this young primate, which indicated that this individual died when it was only 16 months old. The teeth also demonstrated that the toddler, whose gender is difficult to determine because of its age, belonged to a new species, called Nyanzapithecus alesi. The name Alesi comes from the Turkana word “ales,” which means ancestor.
Tafforeau said the thickness of the tooth enamel suggest a classic hominoid diet, which would be similar to that of a modern gibbon, and would consist mostly of fruits and leaves. Researchers estimate that an adult of this species would weigh about 20 pounds.
Turning their attention to the fantastic creature’s ears, the researchers found that it didn’t have a balance organ. That means it couldn’t move as rapidly through trees as a gibbon. The ears of this primate, however, did have fully developed bony ear tubes. These ear structures “absolutely confirmed that these were apes,” said Miller. “We had no specimens between 15 million and 10 million years ago.”

Scientists generally believe apes and humans diverged in their evolution about 7 million years ago. That means this toddler ape belongs to a species that is likely a common ancestor for other apes and humans.
Anthropologist Meave Leakey, a research professor in the Department of Anthropology and the Turkana Basin Institute, suggested that this fossil “gives us a picture for the first time of what the ancestor of apes and humans looked like 13 million years ago. It also suggests,” she continued in an email, “that the nyanzapiehecines were close to the origin of all living apes and humans.”
Leakey described the fossil as one of the most complete skulls of an ape ever found anywhere and indicated it was of an age that is poorly represented in the African fossil record.
The three years between the discovery of the fossil and its unveiling to the world in the Nature article is “actually very quick,” Leakey explained. The images captured through the synchrotron provide detailed pictures of structures that would otherwise be hidden by bone.
Gathering and interpreting these images meant traveling to Grenoble, which, she explained, “takes considerable time.”
Researchers involved in this study said this is just the beginning of the work they will conduct on this rare and detailed fossil. Nengo said they had already collected two terabytes worth of data from their scans. Much of the further study of this ape will involve a closer examination of all of that data.
“A paper coming out in Nature makes it seem like the end of the process,” Miller said. “This is just the beginning.” He is intrigued to learn more about the organization of the brain.
Nengo hopes to bring together researchers for a two- or three-day workshop in September or October at Stony Brook University to tackle the next phase of analysis for Alesi.
As it turns out, September will likely become an important anniversary for Nengo, as he recalls the memory of a day three years ago that didn’t start out particularly well, but that ended with a rare and thrilling fossil find.
Nengo recalled how excited he was to return to the Turkana Basin Institute to show Richard Leakey, the founder of the site, Meave Leakey and Lawrence Martin, the director of TBI. “I had photos on my iPad and they were absolutely thrilled,” said Nengo. “Everybody was beginning the guesswork of wondering what it is.”